In the first part of my series, LabVIEW Data Storage: Overview of TDMS, I introduced TDMS as our preferred file format and pointed users toward exploring and using TDMS themselves. In this post, I'm assuming that you are comfortable handling channels, properties, and data and want to learn more about optimizing your TDMS files to both decrease disk space and improve performance when opening and modifying files.
Preventing File Fragmentation
The TDMS file format is optimized for streaming. Therefore, data is not arranged neatly in the file. Instead, every time data is written to the file, a new chunk is added that includes some header information and then a chunk of binary data. Essentially, the header is specifying what channels and how many samples are represented in the chunk of binary data. This header plus binary data is then just tacked onto the end of the file. This allows an instance where we may have only written 100 samples to one channel, then we switch and write to another channel, and then we come back and write to the first. In a more traditional file format, the entire file would need to be re-organized on disk in order to keep the data sequential. TDMS handles this by using its headers and simply writing sequentially.
You may have noticed that as you write to a TDMS file, two files are created and grow larger as more data is written. The first is the *.tdms file as expected, but the other file is *.tdms_index. The normal *.tdms file is a dump of the packets we discussed above. However, in order to quickly read these packets later, a second file is created that lists each data chunk and exactly where it can be found. If the tdms file is an encyclopedia of data, then the tdms_index file is a table of contents that allows us to flip to the correct page without reading or caring about the pages before it.
I'm simplifying everything a bit, but for our purposes, this is a fair description of how the actual file is laid out. If you're interested, the full details of the on-disk format are available from NI here: TDMS File Format Internal Structure
So what does all this pushing to disk mean to us? Well, assume we are only saving one single sample point at a time to our tdms file. Each time we wrote we would need to generate a large header and write it to disk just to record a few bytes of data. Our file would grow incredibly large, especially compared to the actual data contained! This is called fragmentation. Luckily, National Instruments takes care of this for us in the background. A buffer is created by default for every channel we create and write to. Unfortunately, documentation on this feature is fairly sparse, but based on my testing the default value seems to be one, which is effectively no buffering. This means that each TDMS Write creates a new chunk instead of samples being stored up in the background and then written as one larger chunk that contains samples from multiple TDMS Write calls.
By default, this is usually okay since most applications are only reading, and thus writing, data at a relatively slow rate. However, over a long test, this can lead to high fragmentation and very large files. These files also generate large tdms_index files since there are so many chunks to keep track of. Other than higher disk usage, there is another detriment to these files - it takes much longer to open them later since the entire index needs to be read in and processed. In fact, a good measure of fragmentation is comparing the size of your tdms file to the size of your tdms_index file. The worst-case scenario would be if these files are near identical sizes.
There are two solutions to this problem. The simplest is to run a "defragment" on your tdms file after you are finished generating it. The TDMS Defragment function can be found on the palette with the other TDMS tools. The other solution is to prevent this fragmentation in the first place. You may already be thinking about storing your data in an array and waiting for it to reach a specific size, or maybe just not acquiring data as quickly, but there is a much more elegant solution. NI gives us access to a fairly undocumented property that will allow us to adjust their background buffer. It's called NI_MinimumBufferSize and needs to be specified for each channel you create. There is some documentation from the LabVIEW Help here: Setting the Minimum Buffer Size for a .tdms File.
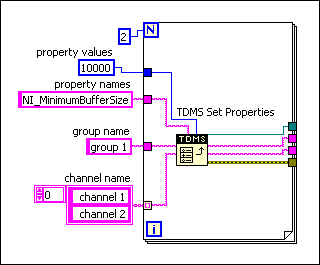
Determining an appropriate size for your application basically involves a balancing act between disk usage, RAM usage, and data integrity. If your application or OS crashed before a buffer is written to disk, your unwritten data will be lost. This can be avoided by using the Flush TDMS function, also available on the TDMS Palette. This function explicitly writes the buffers to disk for a given TDMS file, no matter how many samples are in them. At DMC we often use the Flush TDMS function when changing between test steps, or at a given interval (say 15 minutes) if the data rate is fluctuating or under user control. Obviously, this is very dependent on the specific implementation.
The overall goal here is to minimize our fragmentation in order to keep disk usage low and more importantly to keep opening and interacting with TDMS files quick.
Efficiently Managing Metadata
Remember earlier when I discussed the difference between binary data and metadata such as properties and names? Well, metadata can also have a strong effect on the performance of our TDMS files. Metadata is not stored and opened the same way as the raw binary data. Because metadata includes a lot of the important hierarchy and channel properties for a TDMS file it needs to be quickly accessed. If we go back to the encyclopedia analogy from above, this metadata makes up our table of contents. It would be impossible to look up the data for a specific channel without knowing the names of the channels that exist in our file. This can also be true of other properties, like channel length or datatype. For this reason, all metadata is loaded into memory when you open a TDMS file. It also means that as you write more metadata, your memory footprint grows.
This may seem fairly trivial right now, but consider the following case. You're writing an application that goes through a series of test steps. During the test, you need to monitor data from 500 channels. Maybe some of these are analog inputs, maybe some of them are data values received over CAN or serial, but the application has 500 distinct values that need to be recorded during the test. The simplest implementation is to create a new group for each step the test runs through and then save each of your channels by name into that group. Well, if we estimate an average of 10 letters per channel name, with 500 channels, and 100 steps you now have 500k bytes of extra metadata (at one byte per character). This doesn't take into account the default properties that each channel has, like NI_ChannelLength or NI_DataType. Those account for another 27 characters per channel and another 1350k bytes. When you consider parsing time and reading all of this from different places in disk, your TDMS files will open very slowly indeed, even if the files themselves aren't large.
There are a number of ways to get around this, and I'll talk about them more in depth in my next post.
Reading Only What You Need
Another option for TDMS optimization that is often under-utilized is the ability to only read certain chunks of the raw data. For example, if I am only interested in the last half hour of a test, there is no need to read in the entire data structure and then discard half of it. Instead, I can read the properties of the channel, including length, time interval, and start time and then calculate where I need to start my read and how many samples to retrieve. This doesn't provide a huge benefit with lower data rates or low channel count applications, but if we are talking about an incredibly large TDMS file, then it may not even be possible to pull the full waveform into memory.
This can also be used when analyzing past data. If you are looking for a signal to transition from 0 volts to 10 volts in a larger dataset, the most efficient way to search is via chunking. Chunking is the process of reading in a smaller sample set over and over. Instead of reading in a full array of 100k samples, we can read in 1k samples at a time and check for the voltage transition. If we really wanted to optimize the process we would tune the chunk size (multiples of 2 work well - 1024 is often a good choice) and make sure that we reused the same chunk of data for each read and inspection to prevent multiple memory allocations.
Conclusion
This should provide a good primer on dealing with complicated and larger datasets using the TDMS format. By utilizing the above techniques you can not only substantially decrease the file size, but also increase the performance and responsiveness of opening and managing these files. Obviously, the exact implementation will vary greatly from one application to another, but generally, each of these tips can be used in some facet.
For a hands-on demonstration of how we pulled all of these techniques together to implement a data file format for a Battery Management System (BMS) Validation Test Stand check out my next blog in this series: LabVIEW Data Storage: TDMS Usage Case Study
Learn more about DMC's Data Analysis, Data Mining, and Reporting expertise.